Could all the data produced by YouTube be stored in the volume of a single teaspoon? And could thousands of copies of the Bible be preserved in a unit that is a billionth of a gram? Israeli researchers say, theoretically, yes. And the way to do it is by using DNA for data storage.
In a new study, Israeli researchers at the Technion-Israel Institute of Technology in Haifa and the Interdisciplinary Center (IDC) Herzliya make a case for using DNA as part of a viable, long-term solution to storing our digital libraries as well as the incredible amount of data created every day by humans and machines. And they say they’ve developed a new DNA coding method to do so.
“The density and long-term stability of DNA make it an appealing storage medium, particularly for long-term data archiving,” the researchers say in a study titled “Data storage in DNA with fewer synthesis cycles using composite DNA letters,” published this week in the peer-reviewed scientific journal Nature Biotechnology.
The study was led by research student Leon Anavy at the Technion Faculty of Computer Science under the guidance of Professor Zohar Yakhini of the Technion Faculty of Computer Science and the Efi Arazi School of Computer Science at the Interdisciplinary Center Herzliya (IDC), in collaboration with Professor Roee Amit’s Synthetic Biology Laboratory at the Technion Faculty of Biotechnology and Food Engineering, Dr. Orna Atar, lab manager at the Synthetic Biology Laboratory, and research student Inbal Vaknin.

“DNA is what every organism uses to store its most valuable information, which is its genetic information,” Anavy tells NoCamels in a phone interview. “Evolution has made DNA an ideal media for storing information; we are simply trying to copy what nature has already been doing.”
“In nature, DNA stores genetic information for billions of years and has evolved to be a robust and efficient mechanism for carrying and handling information,” Avany writes in a blog post on the study published on Tuesday.
In addition, Anavy and the researcher team say the new encoding and decoding methods they developed at the Technion’s Genome Center allows for longer-term (thousand-fold) retention of information on DNA, a faster storage process, and zero energy and maintenance costs.
The Data Era and the rise of DNA storage
The speed with which data is created quickly outpaces the current abilities to process and store it, creating a uniquely 21st-century problem.
And just how much data is out there?
By 2020, it is estimated that for every person on the planet, 1.7MB of data will be created every single second, according to a 2018 Data Never Sleeps report which tracks the data explosion worldwide. Ninety percent of all data today was created just since 2015, accounting for some 2.5 quintillion bytes of data per day, according to a previous report.
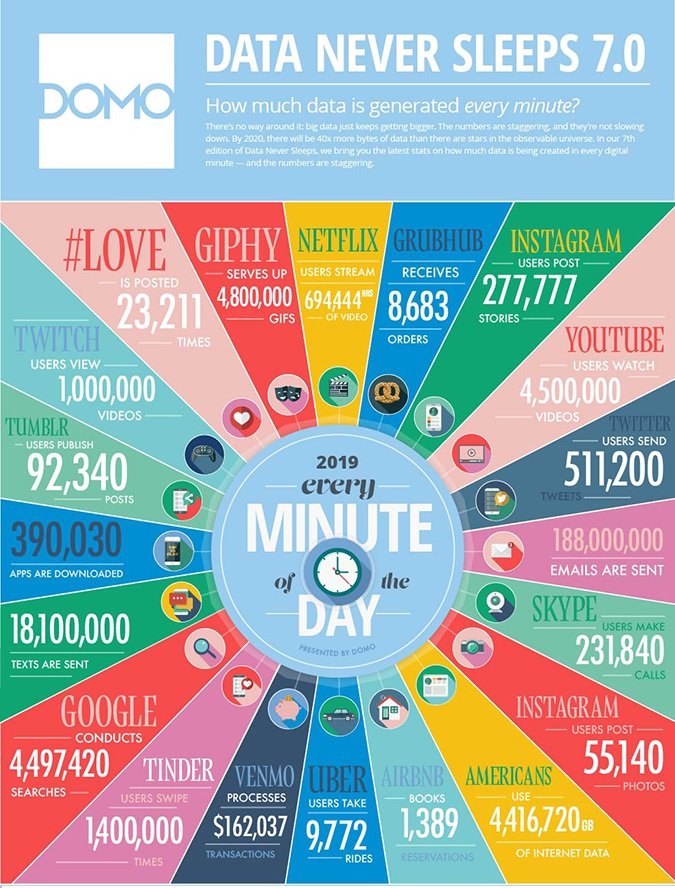
And by next year, there will be 40 times more bytes of data in the world than there are stars in the “observable universe,” says the most recent Data New Sleeps report. For every minute in 2019, Instagram users post 277,777 stories and over 50,000 photos, 188 million emails are sent, Google conducts over 4.4 million searches, and Netflix users stream close to 700,000 hours of video. For every minute. These numbers are staggering.
The current infrastructure can only handle some of the data.
According to US startup Catalog, which says it is building the world’s first DNA-based platform for massive digital data storage and computation, the world is set to generate 160 zettabytes of data in 2025.
“Conventional storage media like flash-drives and hard-drives do not have the longevity, data density, or cost efficiency to meet the global demand,” the company says. Hard discs and memory chips may still be around but the use of the tech is all but obsolete and will likely not be around 20 years from now (we’re looking at you, floppy discs).
Magnetic tape, which is cheap but takes up physical space, is used to store “most digital archives — from music to satellite images to research files –according to a 2018 WIRED piece. And cloud environments require enormous server farms that consume vast amounts of resources including energy and land even as they are a popular solution.

The idea of storing huge amounts of data in DNA has gained momentum in recent years. It began sometime in 2011 when British bioinformaticists were discussing how they could conceivably store the reams of genome sequences and other data coming in at break-neck speed, according to a 2016 report in Nature. (By 2025, between 100 million and 2 billion human genomes could have been sequenced, according to a 2015 study.)
The scientists joked that they could use the actual DNA for the storage, which one of them described as a “lightbulb moment.”
In 2013, these same scientists announced that they had successfully used DNA to encode five files at 739 KB including all 154 of Shakespeare’s sonnets and a snippet of Martin Luther King’s “I have a dream” speech. This was preceded by the work of an American biologist who encoded the draft of a whole book containing 53,246 words, 11 image files and a JavaScript program into DNA.
This summer, Catalog announced that it successfully stored all 16GB of Wikipedia’s English-language text on tiny DNA strands in a laboratory vial.
Sign up for our free weekly newsletter
SubscribeThe technology for storing information on DNA clearly exists, but there are a host of challenges ahead to using it as a viable alternative to existing digital data solutions, “from reliably encoding information in DNA and retrieving only the information a user needs, to making nucleotide [a building block of DNA] strings cheaply and quickly enough,” according to Nature.
The use of composite DNA letters
In their latest study, the Israeli researchers say they built upon a redundancy in existing DNA storage technologies to demonstrate the storage of information in a density of more than 10 petabytes (one petabyte is one million gigabytes) in a single gram of synthetic DNA, while significantly improving the writing process.
“Storing digital information on DNA involves encoding the information into a sequence over the DNA alphabet (that is, A, C, G and T), producing synthetic DNA molecules with the desired sequence and storing the synthetic biological material,” the researchers wrote in their study. “Reading the stored information requires sequencing of the DNA and decoding to obtain the original digital information.”
Using composite DNA letters (a representation of a position in a sequence that consists of a mixture of all four DNA nucleotides in a predetermined ratio), the researchers said they increased the effective number of letters beyond the four building blocks in natural DNA, using new letters that are unique combinations of the original letters and allowing more information to be encoded in each letter in the sequence.
The idea is similar to the formation of new colors using mixtures of base colors, Anavy explains to NoCamels.
“Let’s say you’re an artist and you only have these three base colors: yellow, red, and blue. Now the standard way is to only paint with these three colors. What we are saying is that maybe we can mix them up and create new colors for a richer palette to paint with,” he says.
Generating this richer alphabet for DNA encoding is like mixing these base colors, he concludes.

Professor Yakhini explained further in the Technion statement that “the current synthesis and sequencing processes are inherently redundant because each molecule is produced in large numbers and is read in multiple copies during sequencing.”
“The method we developed leverages this redundancy to increase the effective number of letters well over the original four letters, making it possible for us to encode and write each unit of information in fewer cycles of synthesis,” he added.
The team said its work showed a reduction of the number of synthesis rounds required per unit of information by 20 percent, with a potential future reduction of up to 75 percent. This means a faster, affordable storage process.
“In this work, we have implemented a DNA based storage system that encodes information with synthesis efficiency that is significantly better than the standard approach,” explained Professor Amit in the statement. “The study included the actual implementation of the new coding technique for storing large-volume information on DNA molecules and reconstructing it for testing the process.”
Using these methods, a test tube containing about 10 nanograms (billionths of a gram) of DNA encoded with thousands of copies of a bilingual version of the Bible, was produced and currently sits on one of the shelves in Prof. Amit’s lab at the Technion, the university said.
The researchers also developed an advanced error-correction mechanism to the storage process in order to minimize errors.
“When working in a system consisting of millions of parts (molecules), even one-in-a-million events occur, which can disrupt the reading,” Anavy said in the university statement.
“Thanks to the use of error-correction codes that are tailored to the unique encoding we created, we were able to perform highly efficient coding and to successfully recover the information,” he added.
The researchers say that the technology they presented in their study could potentially further streamline processes in synthetic biology and biotechnology.
“We believe that in the coming years, we will see a significant increase in the use of synthetic DNA in research and industry,” they concluded.
Full disclosure: The IDC is a sponsor of NoCamels
Related posts
Editors’ & Readers’ Choice: 10 Favorite NoCamels Articles

Forward Facing: What Does The Future Hold For Israeli High-Tech?

Impact Innovation: Israeli Startups That Could Shape Our Future




Facebook comments